Multi-organ detection method in abdominal CT images based on deep differentiable random forest
-
摘要:
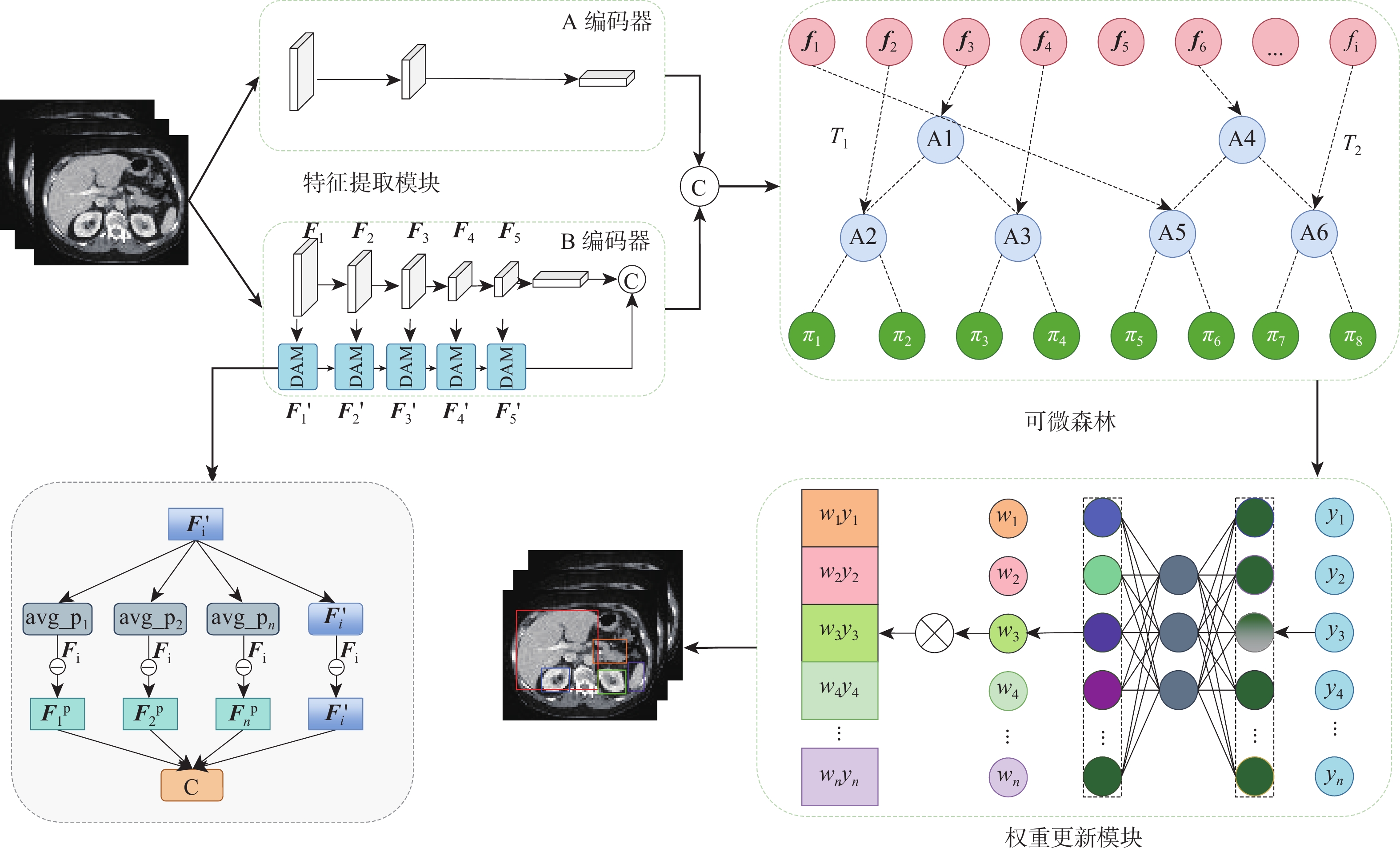
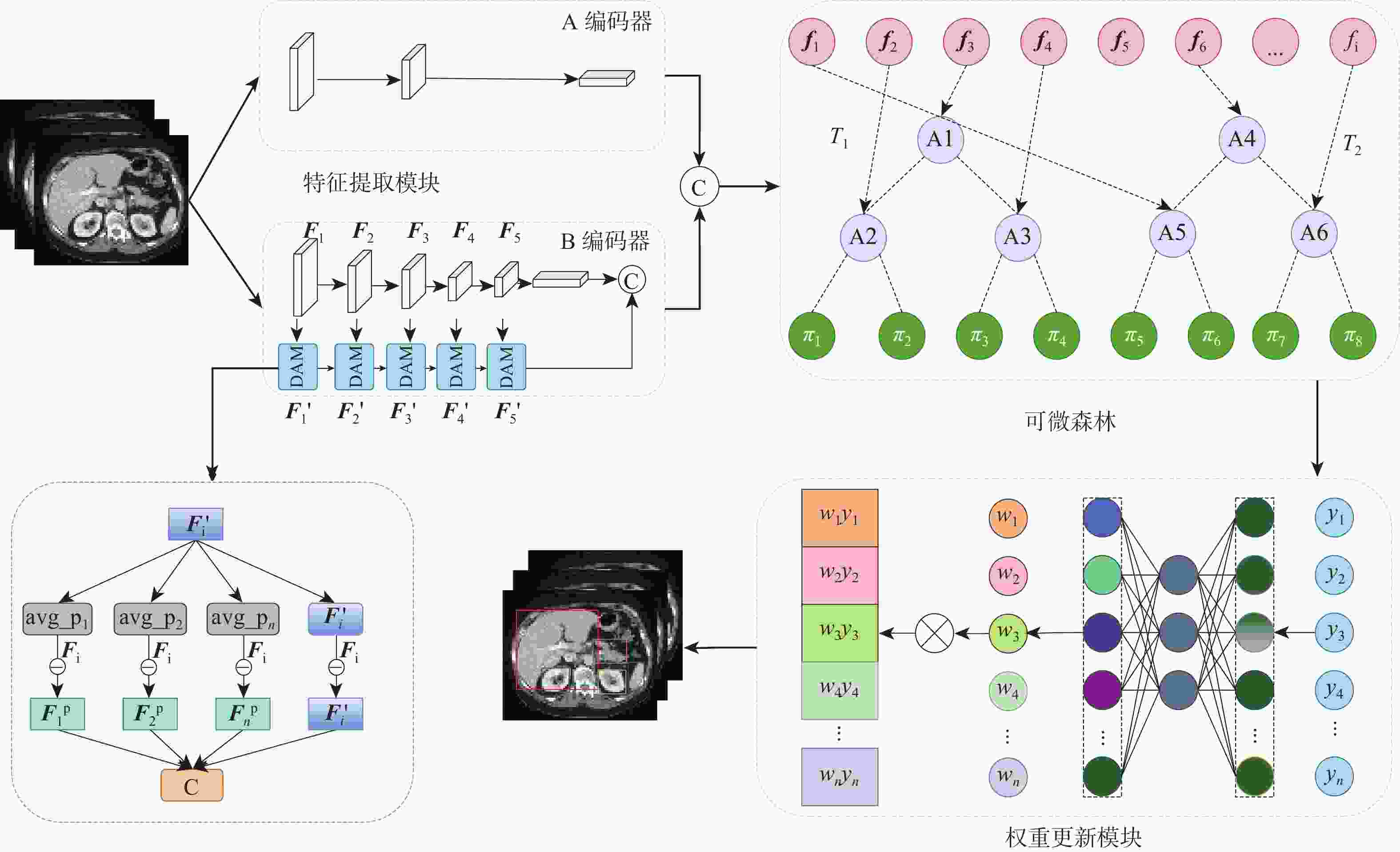
针对传统随机森林方法在处理高维、结构复杂医学图像数据特征学习不充分问题,提出一种基于深度可微随机森林的腹部多器官检测方法。所提方法先将深度学习和随机森林进行巧妙地融合,使用全局-局部双编码器提取高层特征,再将特征输入到可微随机森林进行树分裂。所设计的决策注意力为每棵决策树分配权重,利用反向传播学习参数,构造最终的端到端的检测模型。与传统随机森林不同的是,所提方法采用概率的形式进行树节点分裂,多棵决策树以参数权重进行投票。这种反向传播学习节点分裂参数和投票权重参数策略,可以避免传统随机森林叶节点分裂造成的局部最优,使深度可微随机森林能寻找出全局最优值。在2个公共医学图像多器官数据集(AbdomenCT-1K、AMOS2022)上进行了实验,在AbdomenCT-1K中5个腹部器官的平均WD值比对比方法减少了0.7~2.66 mm,在AMOS2022中7个腹部器官的平均WD值比对比方法减少了0.67~2.68 mm。结果表明:所提方法具有较高的检测准确性。
Abstract:To address the insufficient feature learning of traditional random forests when processing high-dimensional and structurally complex medical images, this paper proposes an abdominal multi-organ detection method utilizing deep differentiable random forests methdo. This method first ingeniously fused deep learning with random forest, employing a global-local dual encoder to extract high-level features that were subsequently fed into a differentiable random forest for tree partitioning. The designed decision attention assigned weights to each decision tree, and all parameters were learned through backpropagation to construct the final end-to-end model. Unlike traditional random forests, this method performs tree node splitting in the form of probabilities, and multiple decision trees vote with weighted parameters. This backpropagation learning of node splitting parameters and voting weight parameters can avoid the local optimum brought by traditional random forest leaf node splitting, allowing the deep differentiable random forest to search for the global optimum. Ultimately, experiments were carried out on two public medical image multi-organ datasets (AbdomenCT-1K and AMOS2022). The findings reveal that, in comparison to the benchmark method, the average WD value for five organs in AbdomenCT-1K decreases by 0.7−2.66 mm, while the average WD value for seven organs in AMOS2022 declines by 0.67−2.68 mm. These demonstrate that the proposed method achieves superior detection accuracy.
-
表 1 AbdomenCT-1K数据集对比实验(WD)
Table 1. Comparative experiments on the AbdomenCT-1K dataset (WD)
表 2 AMOS2022数据集对比实验(WD)
Table 2. Comparative experiments on the AMOS2022 dataset (WD)
表 3 AbdomenCT-1K消融实验结果(WD)
Table 3. Results of ablation experiments on AbdomenCT-1K dataset (WD)
方法 平均WD值 肝脏 脾脏 胰腺 左肾 右肾 无精细特征 7.4 5.6 8.7 5.2 4.6 无权重学习 7.6 6.1 8.5 5.6 5.1 本文方法 6.5 5.3 8.2 4.1 3.8 表 4 AMOS2022消融实验结果(WD)
Table 4. Results of ablation experiments on AMOS2022 dataset (WD)
方法 平均WD值 肝脏 脾脏 胰腺 左肾 右肾 胆囊 膀胱 无精细特征 7.7 6.7 9.3 5.1 5.1 9.2 9.3 无权重学习 7.8 6.9 9.2 5.2 5.3 9.3 9.4 本文方法 7.2 6.5 8.9 4.8 4.7 8.7 9.1 -
[1] GOLDMAN L W. Principles of CT and CT technology[J]. Journal of Nuclear Medicine Technology, 2007, 35(3): 115-128. doi: 10.2967/jnmt.107.042978 [2] WANG Y, ZHANG Y, LIU L, et al. FGB: feature guidance branch for organ detection in medical images[C]//Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging. Piscataway: IEEE Press, 2020: 349-353. [3] XUN S, LI D, ZHU H, et al. Generative adversarial networks in medical image segmentation: a review[J]. Computers in Biology and Medicine, 2022, 140: 105063. doi: 10.1016/j.compbiomed.2021.105063 [4] WIERZBICKI R, PAWLOWICZ M, JOB J, et al. 3D mixed-reality visualization of medical imaging data as a supporting tool for innovative, minimally invasive surgery for gastrointestinal tumors and systemic treatment as a new path in personalized treatment of advanced cancer diseases[J]. Journal of Cancer Research and Clinical Oncology, 2022, 148(1): 237-243. doi: 10.1007/s00432-021-03680-w [5] FENCHEL M, THESEN S, SCHILLING A. Automatic labeling of anatomical structures in MR FastView images using a statistical atlas[C]//Proceedings of the Medical Image Computing and Computer-Assisted Intervention. Berlin: Springer, 2008: 576-584. [6] CRIMINISI A, ROBERTSON D, KONUKOGLU E, et al. Regression forests for efficient anatomy detection and localization in computed tomography scans[J]. Medical Image Analysis, 2013, 17(8): 1293-1303. doi: 10.1016/j.media.2013.01.001 [7] GAURIAU R, CUINGNET R, LESAGE D, et al. Multi-organ localization with cascaded global-to-local regression and shape prior[J]. Medical Image Analysis, 2015, 23(1): 70-83. doi: 10.1016/j.media.2015.04.007 [8] DE VOS B D, WOLTERINK J M, DE JONG P A, et al. 2D image classification for 3D anatomy localization: employing deep convolutional neural networks[C]//Proceedings of the SPIE Medical imaging 2016. San Diego: SPIE, 2016, 9784: 517-523. [9] HUMPIRE-MAMANI G E, SETIO A A A, VAN GINNEKEN B, et al. Efficient organ localization using multi-label convolutional neural networks in thorax-abdomen CT scans[J]. Physics in Medicine & Biology, 2018, 63(8): 085003. [10] XU X, ZHOU F, LIU B, et al. Efficient multiple organ localization in CT image using 3D region proposal network[J]. IEEE Transactions on Medical Imaging, 2019, 38(8): 1885-1898. doi: 10.1109/TMI.2019.2894854 [11] 施俊, 汪琳琳, 王珊珊, 等. 深度学习在医学影像中的应用综述[J]. 中国图象图形学报, 2020, 25(10): 1953-1981. doi: 10.11834/jig.200255SHI J, WANG L L, WANG S S, et al. Applications of deep learning in medical imaging: a survey[J]. Journal of Image and Graphics, 2020, 25(10): 1953-1981(in Chinese). doi: 10.11834/jig.200255 [12] BREIMAN L. Random forests[J]. Machine Learning, 2001, 45: 5-32. doi: 10.1023/A:1010933404324 [13] BIAU G. Analysis of a random forests model[J]. The Journal of Machine Learning Research, 2012, 13: 1063-1095. [14] 邓浩伟, 侯月皎, 张朝月, 等. 基于级联森林和多模态融合的脑力疲劳识别算法[J]. 北京航空航天大学学报. 2025, 51(2): 584-593.DENG H W, HOU Y J, ZHANG C Y, et al. Mental fatigue recognition algorithm based on cascade forest and multi-modal fusion[J]. Journal of Beijing University of Aeronautics and Astronautics, 2025, 51(2): 584-593(in Chinese). [15] 杨晔民, 张慧军, 张小龙. 随机森林的可解释性可视分析方法研究[J]. 计算机工程与应用, 2021, 57(6): 168-175.YANG Y M, ZHANG H J, ZHANG X L. Research on interpretable visual analysis method of random forest[J]. Computer Engineering and Applications, 2021, 57(6): 168-175(in Chinese). [16] SILVA A, GOMBOLAY M, KILLIAN T, et al. Optimization methods for interpretable differentiable decision trees applied to reinforcement learning[C]//Proceedings of the International Conference on Artificial Intelligence and Statistics. NewYork: PMLR, 2020: 1855-1865. [17] LI Z, LIU F, YANG W, et al. A survey of convolutional neural networks: analysis, applications, and prospects[J]. IEEE Transactions on Neural Networks and Learning Systems, 2021, 33(12): 6999-7019. [18] CRIMINISI A, ROBERTSON D, PAULY O, et al. Anatomy detection and localization in 3D medical images[J]. Decision Forests for Computer Vision and Medical Image Analysis, 2013, 193-209. [19] MATSUO Y, LECUN Y, AHANI M, et al. Deep learning, reinforcement learning, and world models[J]. Neural Networks, 2022: 267-275. [20] CHEN Y, CHI Y, FAN J, et al. Gradient descent with random initialization: fast global convergence for nonconvex phase retrieval[J]. Mathematical Programming, 2019, 176: 5-37. doi: 10.1007/s10107-019-01363-6 [21] WU Z, SHEN C, VAN DEN HENGEL A. Wider or deeper: revisiting the resnet model for visual recognition[J]. Pattern Recognition, 2019, 90: 119-133. doi: 10.1016/j.patcog.2019.01.006 [22] ZHU X, WANG X, SHI Y, et al. Channel-wise attention mechanism in the 3D convolutional network for lung nodule detection[J]. Electronics, 2022, 11(10): 1600. [23] MA J, ZHANG Y, GU S, et al. Abdomenct-1k: is abdominal organ segmentation a solved problem?[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2021, 44(10): 6695-6714. [24] KONDO S, KASAI S, XING X. Multi-modality abdominal multi-Organ segmentation with deep supervised 3D segmentation model[EB/OL]. (2022-08-24)[2023-10-03]. https://doi.org/10.48550/arXiv.2208.12041. [25] YUAN Z, CHEN T, XING X, et al. BM3D denoising for a cluster-analysis-based multibaseline InSAR phase-unwrapping method[J]. Remote Sensing, 2022, 14(8): 1836. doi: 10.3390/rs14081836 [26] PARIS S, KORNPROBST P, TUMBLIN J, et al. Bilateral filtering: theory and applications[J]. Foundations and Trends® in Computer Graphics and Vision, 2009, 4(1): 1-73. [27] HINTZE J L, NELSON R D. Violin plots: a box plot-density trace synergism[J]. The American Statistician, 1998, 52(2): 181-184. doi: 10.1080/00031305.1998.10480559 -





 下载:
下载: